From hidden data and information towards data-driven research
Lisa Langnickel, ZB MED, Cologne
Machine readability and access to data, information and knowledge are core requirements for data-driven research. Furthermore, the enormous growth in freely available, electronic research data increases the need for semantic interoperability as well as computational methods to generate new information and knowledge from the data.
This, however, implies that all published data is stored in a machine-readable format and that data can be accessed. Especially in the medical area, this is hampered by the heterogeneity and missing standardization of the data as well as the restricted access and availability of high quality data.
Concerning the literature, there is an increasing need for text mining solutions to make a transfer from unstructured text to machine readable information possible. During the past 20-30 years, intense research has been done in the field of natural language processing (NLP) and also in the specific application field of bio-medical NLP (bioNLP). Currently, Artificial Intelligence (AI) methods seem to be superior to traditional approaches. The success of those methods is, however, dependent on high-quality, labeled data whose availability is strongly limited. In addition, it still remains open whether the results achieved on the specific training/test data are transferable to real world applications.
Another obstacle for data analysis in the medical domain is the access to personalized health data. Despite the growing amount of freely available data, personal data (e.g. clinical or epidemiological data) is usually not publicly available due to data privacy. To circumvent data protection, machine learning (ML) methods will be investigated in order to generate synthetic data.
The overarching aim of this project is to investigate computational methods in order to make biomedical data and information available in a machine-readable format and, thereby, supporting researchers.
Two examples
Investigating the Robustness of Transfer learning-based NER methods

Figure 1: Excerpt of an abstract (doi: 10.1101/2021.07.06.21260115), annotated with disease mentions (screenshot taken from https://preview.zbmed.de).
We investigated the robustness of current state-of-the-art text mining methods, such as BioBERT [1], in the area of Named Entity Recognition (NER) of diseases. These machine learning (ML)-based methods are usually trained and evaluated on specific, relatively small corpora and evaluations on corresponding test sets show promising results. For NER of diseases, two different manually labeled data sets are publicly available which consist of training, development and test data. Our first investigation focused on cross-corpora evaluation: training on one dataset and evaluation on the test set of the other data set. We could show that the model achieves an f1-score of only 68% - a drop of about 20% compared to the original test set [2]. Provided ML-based models are able to generalize, comparable results would be expected from data sets following the same annotation guidelines. An analysis of the two different data sets revealed that the training and corresponding test set (belonging to the same data set) are similar in wording and topics while the data sets as a whole do not. This leads to the assumption that a model trained on one available corpus is not applicable to real world cases and needs to be continuously retrained (called continual learning). Currently, we are investigating compute- and resource-efficient methods.
Service Science - COVID-19 underlines the need for text mining-based solutions
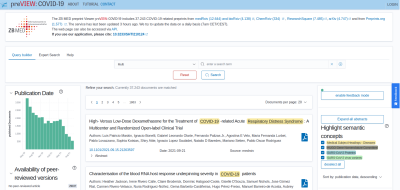
Figure 2: Screenshot of our semantic search engine preVIEW, freely accessible under https://preview.zbmed.de.
The current COVID-19 pandemic underlines the need for text mining methods as more than 100 papers – mostly in form of preprints – are currently published per day which makes it infeasible for a human to read all of them. In order to support researchers to cope with this huge amount of information, we set up a text mining-based semantic search engine, called preVIEW, that currently contains more than 36,000 preprints from seven different preprint servers, such as bioRxiv and medRxiv [3]. In accordance to our previous research, we found out that the current machine learning-based state-of-the-art methods are not applicable to services/real world cases because they do not generalize well and are not consistently able to recognize new terms. For example, for the recognition of diseases, the ML-based algorithm TaggerOne [4] missed new terms like COVID-19. As text mining is nevertheless needed to index these high amounts of preprints and thereby find relevant literature, we extended the text mining workflow with additional rule based components and re-evaluated the resulting annotations. Moreover, for new entity classes – i.e. SARS-CoV-2 specific virus proteins and variants of interest – a dictionary-based approach was implemented due to the lack of training data for supervised learning algorithms.
Whereas preVIEW was developed as a fast prototype together with the user community in the beginning of the crisis, it has been continuously improved towards a sustainable system [5]. In addition, it is currently undergoing evaluation by BioCreative Interactive text mining track [6] in order to evaluate the system usability by a variety of end users.
References
[1] Lee J et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, 2020. DOI:10.1093/bioinformatics/btz682
[2] Langnickel L et al. We Are Not Ready yet: Limitations of Transfer Learning for Disease Named Entity Recognition. bioRxiv, 2021. DOI:10.1101/2021.07.11.451939
[3] Langnickel L et al. COVID-19 preVIEW: Semantic Search to Explore COVID-19 Research Preprints. Public Health and Informatics, 2021. DOI: 10.3233/SHTI210124
[4] Leaman R et al. TaggerOne: joint named entity recognition and normalization with semi-Markov Models. Bioinformatics, 2016. DOI:10.1093/bioinformatics/btw343
[5] Langnickel et al. preVIEW: from a fast prototype towards a sustainable semantic search system for central. Journal of European Association for Health Information and Libraries, 2021. DOI:10.32384/jeahil17484
[6] BioCreative - Track 4- COVID-19 text mining tool interactive demo. Accessed October 5, 2021. https://biocreative.bioinformatics.udel.edu/tasks/biocreative-vii/track-4/




